19 Protein Synthesis II: RNA Processing
Andrea Bierema
Learning Objectives
Students will be able to:
- Describe the molecular anatomy of genes and genomes.
- Describe exons and introns.
- Explain how splicing occurs during mRNA processing.
- Explain the process of alternative splicing.
- Describe how alternative splicing contributes to cells having different functions.
- Identify additional steps that take place during mRNA processing.
Overview
At the end of transcription, once the polymerase releases the RNA molecule, the RNA is not quite ready for translation and is not technically an “mRNA” yet. It still needs to be “processed,” which means that certain nucleotides are removed and others are added—at the end of this, it is an mRNA molecule, but before that, it is a pre-mRNA molecule. This occurs as the RNA molecule is leaving the nucleus. After it is processed, it is then ready for translation, which is covered in a future chapter. This chapter focuses on the different steps that take place during mRNA processing and how some of these steps allow for cells within a single organism to have different functions. Please see the previous chapter for a review on transcription.
Splicing
After an mRNA molecule is produced, specific sequences throughout the mRNA molecule are removed. These sequences are called introns. The sequences that remain are called exons. To recall these terms, consider the following: exons correspond to protein-coding sequences (“EX–on” signifies that they are expressed), and intervening sequences called introns (“IN-tron” denotes their intervening role), which may be involved in gene regulation but are removed from the pre-mRNA during processing. Intron sequences in mRNA do not encode functional proteins.
All of a pre-mRNA’s introns must be completely and precisely removed before protein synthesis. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional. The process of removing introns and reconnecting exons is called splicing.
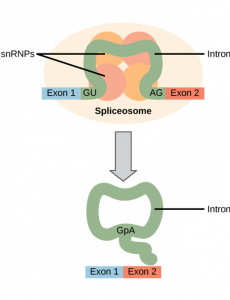
If we consider that a base sequence is similar to letters on a page, then splicing is removing particular lines of text. View the following video, which follows this analogy, for a brief overview of what splicing does (open the summary dialog at the end of the video to reflect on what you learned).
For closed captioning or to view the full transcript see the video on YouTube. Or click on the “YouTube” link in the video.
Exercise
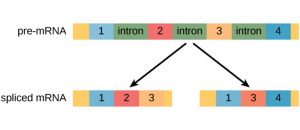
Let’s test your knowledge! Complete the following image by dragging the parts of the pre-RNA that will be in the mature mRNA after mRNA processing. The image includes three exons and two introns. The untranslated regions are already in place.
Alternative Splicing
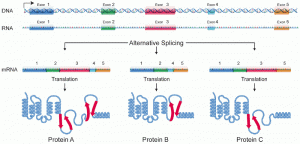
Genes that exhibited alternative RNA splicing were first observed in the 1970s. Alternative RNA splicing is a mechanism that allows different protein products to be produced from one gene when different combinations of exons are combined to form the mRNA. This alternative splicing can be haphazard, but more often it is controlled and acts as a mechanism of gene regulation, with the frequency of different splicing alternatives controlled by the cell as a way to control the production of different protein products in different cells or at different stages of development. Alternative splicing is now understood to be a common mechanism of gene regulation in eukaryotes; according to one estimate, 70% of genes in humans are expressed as multiple proteins through alternative splicing. Although there are multiple ways to alternatively splice RNA transcripts, the original 5′-3′ order of the exons is always conserved. That is, a transcript with exons 1 2 3 4 5 6 7 might be spliced 1 2 4 5 6 7 or 1 2 3 6 7, but never 1 2 5 4 3 6 7.
Here is another representation of alternative splicing—this one does not label the introns (assume the molecule segment between the exons are introns), like the previous but it shows how alternative splicing impacts the protein.
How could alternative splicing evolve? Introns have a beginning—and ending—recognition sequence; it is easy to imagine the failure of the splicing mechanism to identify the end of an intron and instead find the end of the next intron, thus removing two introns and the intervening exon. In fact, there are mechanisms in place to prevent such intron-skipping, but mutations are likely to lead to their failure. Such “mistakes” would more than likely produce a nonfunctional protein. Indeed, the cause of many genetic diseases is abnormal splicing rather than mutations in a coding sequence. However, alternative splicing could possibly create a protein variant without the loss of the original protein, opening up possibilities for adaptation of the new variant to new functions. Gene duplication has played an important role in the evolution of new functions in a similar way by providing genes that may evolve without eliminating the original, functional protein.
Additional Processing
Before the mRNA leaves the nucleus, it is given two protective “caps” that prevent the ends of the strand from degrading during its journey. The two ends of a DNA strand are referred to as 3′ and 5′, which references the position of sugar molecules in the DNA. The 5′ cap is placed on the 5′ end of the mRNA. The poly-A tail, which is attached to the 3′ end, is usually composed of a long chain of adenine (A) nucleotides. These changes protect the two ends of the RNA from being broken down by other enzymes in the cell.
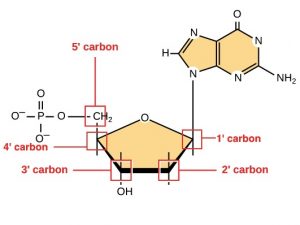
5′ Capping
While the pre-mRNA is still being synthesized, a 7-methylguanosine cap is added to the 5′ end of the growing transcript by a phosphate linkage. This functional group protects the nascent mRNA from degradation. In addition, factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes.
3′ Poly-A Tail
Once elongation is complete, the pre-mRNA is cleaved by an endonuclease between an AAUAAA consensus sequence and a GU-rich sequence, leaving the AAUAAA sequence on the pre-mRNA. An enzyme called poly-A polymerase then adds a string of approximately 200 A residues, called the poly-A tail. This modification further protects the pre-mRNA from degradation and is the binding site for a protein necessary for exporting the processed mRNA to the cytoplasm. Below is a more complete diagram of mRNA processing.

Test Your Understanding
Attribution
This chapter is a modified derivative of the following articles:
“Genes and Proteins” by OpenStax College, Biology 2e, CC BY 4.0. Download the original article for free at https://openstax.org/books/biology-2e/pages/15-introduction
Clark, M. A., Douglas, M., & Choi, J. (2018). Biology 2e. OpenStax. Licensed under CC BY 4.0. Modified by Andrea Bierema.https://openstax.org/books/biology-2e/pages/15-introduction

