21 Protein Synthesis V: Additional Regulation
Andrea Bierema
Learning Objective
Students will be able to:
- Identify ways in which gene expression is regulated during and after translation.
Overview
We have learned how we synthesize proteins and how gene expression is regulated before and after transcription, but the regulation can also happen during or after translation.
Regulation of Translation
We already saw how miRNAs can inhibit translation, but there are a number of other ways that translation of an mRNA can also be regulated in a cell. One key step for regulation is translation initiation.
In order for translation to begin, the ribosome, an RNA-and-protein complex that houses translation, must assemble on the mRNA. This process involves many “helper” proteins, which make sure the ribosome is correctly positioned. Translation can be regulated globally (for every mRNA in the cell) through changes in the availability or activity of the “helper” proteins.
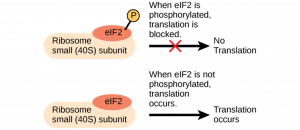
For example, in order for translation to begin, a protein called eukaryotic initiation factor-2 (eIF-2) must bind to a part of the ribosome called the small subunit. Binding of eIF-2 is controlled by phosphorylation, or addition of a phosphate group to the protein.
When eIF-2 is phosphorylated, it’s turned “off”—it undergoes a shape-change and can no longer play its role in initiation, so translation cannot begin. When eIF-2 is not phosphorylated, in contrast, it’s “on” and can carry out its role in initiation, allowing translation to proceed.
Chemical Modifications, Protein Activity, and Longevity
Proteins can be chemically modified with the addition of groups including methyl, phosphate, acetyl, and ubiquitin groups. The addition or removal of these groups from proteins regulates their activity or the length of time they exist in the cell. Sometimes these modifications can regulate where a protein is found in the cell—for example, in the nucleus, in the cytoplasm, or attached to the plasma membrane.
Chemical modifications occur in response to external stimuli such as stress, the lack of nutrients, heat, or ultraviolet light exposure. These changes can alter epigenetic accessibility, transcription, mRNA stability, or translation—all resulting in changes in the expression of various genes. This is an efficient way for the cell to rapidly change the levels of specific proteins in response to the environment. Because proteins are involved in every stage of gene regulation, the phosphorylation of a protein (depending on the protein that is modified) can: alter accessibility to the chromosome; alter translation (by altering transcription factor binding or function); change nuclear shuttling (by influencing modifications to the nuclear pore complex); alter RNA stability (by binding or not binding to the RNA to regulate its stability); modify translation (increase or decrease); or change post-translational modifications (add or remove phosphates or other chemical modifications).
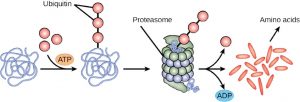
The addition of a ubiquitin group to a protein marks that protein for degradation. Ubiquitin acts like a flag indicating that the protein lifespan is complete. These proteins are moved to the proteasome, an organelle that functions to remove proteins, to be degraded. One way to control gene expression, therefore, is to alter the longevity of the protein.
Attribution
This chapter is a modified derivative of “Eukaryotic translational and post-translational gene regulation,” by OpenStax College, Biology 2e, CC BY 4.0. Download the original article for free at https://openstax.org/books/biology-2e/pages/16-6-eukaryotic-translational-and-post-translational-gene-regulation
Media Attributions
- Central Dogma (no protein) © Andrea Bierema is licensed under a CC BY-NC-SA (Attribution NonCommercial ShareAlike) license
- Ribosomal Regulation © OpenStax is licensed under a CC BY (Attribution) license
- Degradation Proteins © OpenStax is licensed under a CC BY (Attribution) license

